Highlights:
- How do download/backup sequencing data from Illumina basespace
- Using Amazon AWS to store and potentially process the sequencing data
Assumptions for this tutorial:
- Amazon AWS account
- Illumina basespace account (containing sequencing data)
DNA sequencing using Illumina SBS technology (NGS) has become one of the most important workflow in modern research but also diagnostics, plant breeding just to name a few. The standard workflow is to sequence the DNA of interesst using a DNA sequencer, which might be either a local benchtop sequencer (MiSeq/NextSeq) or a production-scale sequencer as part of a larger sequencing facility (HiSeq/NovaSeq). Regardless, the results are usually send to the Illumina cloud (aka basespace) even if the sequencer is on site. For on-premise data storage, have a look here [1].
Once the data arrived on basespace, there is a few options on how to process it, either using Illuminas in-house cloud workflows (e.g. Dragen), in which the processing is done within the basespace cloud, or by retrieving the data from basespace in order to process it somewhere else.
I usually found myself requiring the latter (as do many others) for the following reasons:
- backing up the sequencing data
- running a customized workflow which not covered by Dragen
- limited storage
As for the last point, basespace usually produces the following notification:
We’re writing to notify you that your BaseSpace™ Sequence Hub account has over 900 GB of data stored in it. The maximum allowed in a Basic tier account is 1.
Well you could opt for a Pro or Enterprise basespace account, but chances are you want to store it somewhere since it's either cheaper and more accessible. But also as for the 2nd reason, if the basepace tools such as Dragen and the like don't fit your needs (which is often the case), you probably want run some custom pipelines on your data, which requires some sort of data transfer anyway.
Ideally this would also involve not having to copy anything to my local computer, i.e. the data should be transferred between basepace and some sort of cloud service. An obvious choice is to use AWS, since basespace is already hosted on the AWS in the first place.. You'd think such as transfer should be quite straightforward, however there is surprisingly little information to be found on how to pull this off! Given on how common this is ought to be, I was really surprised by the lack of a straightforward simple walk-through that could be easily followed. [2] In fact I didn't even know how to approach this from a high level, i.e. is there a button somewhere on basepace which does all for you or if not which AWS service are needed.. Even the official Illumina guide is not really helpful:
Can BaseSpace files be transferred directly to AWS (Amazon Web Services)? There is no direct BaseSpace-to-S3 (AWS) transfer system. Installing BaseSpace CLI within an S3 bucket is the best solution. See the for information on installing and using the BaseSpace CLI.
Well, at least they give a good direction, but far from a step-by-step guide on how to actually do this.
Although being an experienced bioinformatician, I still felt at lost. Since I'm likely not the only one in this situation, I thought to write this blog on how I do this!
As of above, it seems the best way to catch both flies with one stone is AWS, i.e. storing the sequence data (i.e. on S3) and use the computing power to run your pipelines (i.e. EC2, nextflow ). But AWS can be overwhelming with it's 200+ services, but no worries, we'll need just a few and I show you which.
High level description: Basespace -> AWS
Having done some research I think the ultimate goal is to deposit the raw data (e.g. fastq files) on a S3 bucket. S3 could then serve as long term storage for your data and can be easily accessed by custom pipelines.
The way to do actually do this on a high-level:
- create an EC2 instance
- install basepace-cli on the instance
- add some temporary storage (EBS)
- download the sequencing data (fastq files) on the EBS disk using basespace-cli
- (optional) run some QC or custom analysis on the data
- push data from EBS to S3 bucket (using AWS sync)
All steps assume to have a working AWS setup configured. If not, you can follow this guide.
Create an EC2 instance
Details can be found here, below is a short summary:
- Log in to the AWS Management Console and select EC2 from the list of services.
- Click the “Launch Instance” button.
- Select an Amazon Machine Image (AMI) for your instance.
- The default is fine (Amazon Linux AMI)
- Choose an instance type.
- The free tier (t2.micro) are unfortunatelly not sufficient, the minimum is in my experience t2.small, but would go for t2.medium!
- Configure instance details.
- Add storage to your instance.
- Add tags to your instance.
- Configure security group.
- Review and launch your instance.

Default for most (5-8) of the above should be fine
Create and attach temporary storage (EBS)
The default instance comes with 30GB storage which is nowhere near enough. We need to attach another EBS Volume:
Additional details on creating volumes and attaching volumes on the links. However I found this tutorial
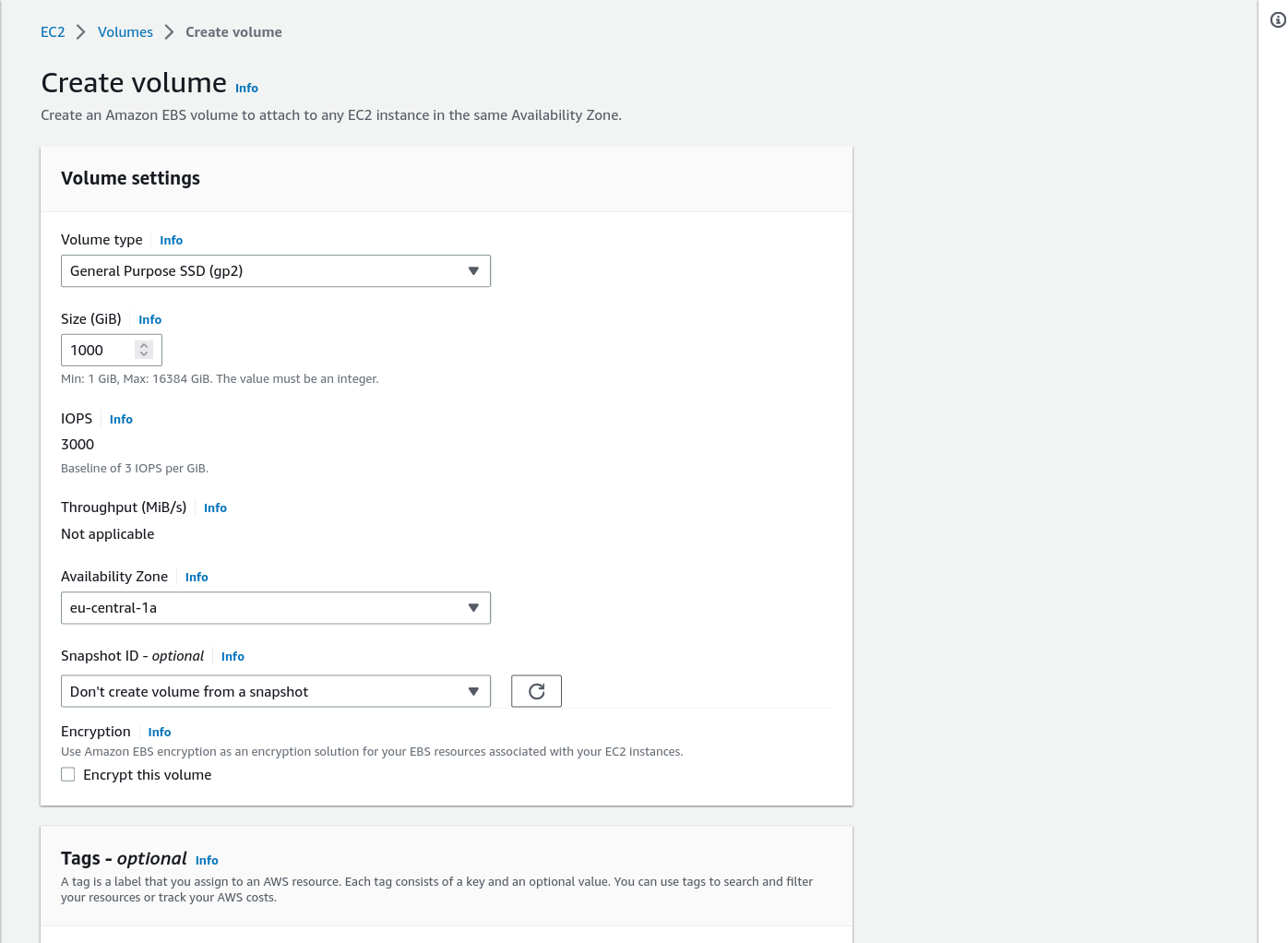 very useful, below the main points:
very useful, below the main points:
- Log in to the AWS Management Console and select EC2 from the list of services.
- Click the “Volumes” under "Elastic Block Store"
- Leave defaults except for Volume size. I'd go for 1000GB which should be enough even for a NovaSeq run.
- ssh in the EC2 instance
- execute the commands below:
$ sudo mkfs -t ext4 /dev/xvdf
mke2fs 1.46.5 (30-Dec-2021)Mount it to a new directory (use any directory name you like):
$ mkdir data
$ sudo mount /dev/xvdf dataBy default, the new directory belongs to the root user. Change the ownership so you don’t need root permission to access it:
$ sudo chown ec2-user dataTest if you can write files into that new disk:
Recall that EBS price is $100/TB/month and S3 price is $23/TB/month
After attaching, the lsblk command will show the new 1000 GB volume.
https://docs.aws.amazon.com/AWSEC2/latest/UserGuide/ebs-using-volumes.html
Note To avoid excessive costs, read the information below:
https://aws.amazon.com/premiumsupport/knowledge-centWer/ebs-charge-stopped-instance/
You are charged for Amazon EBS storage for the amount of storage provisioned to your account, measured in "gigabyte-months." Amazon EC2 instances accrue charges only while they're running. However, EBS volumes attached to instances continue to retain information and accrue charges, even when the instance is stopped
To stop Amazon EBS-related charges, delete EBS volumes that you don't need.
Important: this is probably the biggest factor in terms of costs.
https://www.iobasis.com/Strategies-to-reduce-Amazon-EBS-Storage-Costs/ These are charged by GB provisioned. So the biggest the volume, the more you pay.
They are also charged from the moment they are created until they are eliminated. So it doesn’t matter if it’s not attached to an instance, or it’s attached to a stopped instance. They will be charged anyway.
Another important point is that provisioned means that it doesn’t matter how much data is inside the volume. For example, you provision a 500 GB volume, but you only use 10GB. Then you will be charged for the whole 500 GBs, independently that most volume space is unused.
Using “Delete on Termination”
The Delete on Termination is an option you could set when launching your EC2 instance. It allows you to automatically remove the EBS volume (attached to an EC2 instance) when that instance is terminated. So this avoids keeping an unattached volume. You should use it with caution because data on EBS volume will be lost.
Large files in EBS volumes could also be moved to S3. For example, Standard S3 rates are 77% below General Purpose SSD (gp2) volume prices per GB. Additionally, you pay only for data used, and S3 capacity is doesn’t need to be provisioned. And the data is replicated across three AZs at least.
- Amazon EBS volume types https://docs.aws.amazon.com/AWSEC2/latest/UserGuide/ebs-volume-types.html?icmpid=docs_ec2_console
Install basespace-cli on EC2 instance
Now that we have a functioning "computer", we have can use basespace-cli to download the sequencing data.
There are actually 2 options to download, basespace-cli and basespace mount. I found the cli more robust and reliable, hence will focus on cli here, but will mention a few details for basespace mount further below.
To install the cli, follow this guideline for more details, in short:
mkdir $HOME/bin
wget "https://launch.basespace.illumina.com/CLI/latest/amd64-linux/bs" -O $HOME/bin/bs
chmod +x $HOME/bin/bs
PATH=$PATH:/binAuthenticate Illumina account using below:
bs auth
j# or
bs auth --api-server=https://api.euw2.sh.basespace.illumina.comDownload the sequencing data (fastq files) on the EBS disk using basespace-cli
There are multiple ways to download fastq files, however I found "project" centric download most useful (alternative being runs and session for instance).
First we want to list all possible projects, with even saving them in a csv file:
bs list projects
bs list project -f csv -F Name -F Id -F DateModified -F TotalSize | sed 's/\ /_/g' > myprojects.csvAn example entry might look like this:
Name,Id,DateModified,TotalSize
my_project1,5955954,2023-03-08_15:33:23_+0000_UTC,1564137469
my_project2,5965962,2023-03-08_15:33:23_+0000_UTC,1234899834Now we can perform the actual download, naming the output directory after the project using the following syntax bs download project -i <ID> --extension=fastq.gz -o ~/path/of/your/choice.[^1]
bs project download -i 5955954 -o my_project1 -v --concurrency=low --retry 5This the most important step. This will download all fastq.gz files from your project. Note you have to copy paste the specific ID from your project into this. See the second link below for how to download from multiple projects, subsets of projects, or how to sort by sample names.
[^1]: in order to let the download run in the background, I encourage to use tmux before starting
(optional): Run QC and/or custom analysis on the data
Since the data is downloaded, it is possible to use the instance to run some analysis.
I.e. you could run fastqc as a simple example:
find ./ -name "*.gz" -exec fastqc {} \;Upload to AWS S3:
Create S3 bucket
First we need to create a S3 bucket on AWS as below (using default values).
- Go to AWS management Console and Open S3 Service
- create bucket (bottom)
- Enter a bucket name that is unique. And click on next.
 Here it is named
Here it is named mynewbasespacebucket
Also we need to install AWS cli:
Install AWS cli on EC2
sudo apt install awscli https://docs.aws.amazon.com/cli/latest/userguide/getting-started-quickstart.html
$ aws configure
AWS Access Key ID [None]: AKIAIOSFODNN7EXAMPLE
AWS Secret Access Key [None]: wJalrXUtnFEMI/K7MDENG/bPxRfiCYEXAMPLEKEY
Default region name [None]: us-west-2
Default output format [None]: json
aws s3 ls
2023-02-11 15:45:40 mynewbasespacebucketPush data from EBS to S3 bucket (using AWS sync)
Run the following command to sync the data from the EBS volume to the S3 bucket:
aws s3 sync <ebs_volume_path> s3://<bucket_name>I.e.:
dir=my_project1
aws s3 sync ~/data/${dir} s3://mynewbasespacebucket/${dir}
INFO: console logging re-enabled...
...
my_project1_376308970.json 506 B / 506 B [========================] 100.00% 51.03 KiB/s 0s
INFO: Completed in 13h14m9.869538508s
That's it, the whole project is now sitting in a S3 project. You can delete rm -r my_projects1 the downloaded files now if you want to free space for another project or delete the EBS volume to save costs.
